Electron Microscopy: 3D reconstruction from 2D projections
Bioinformatics
Computational Geometry
Software
Publications
Electron Microscopy: 3D reconstruction from 2D projections
In the hierarchy of biological visualization, 3D Electron Microscopy
(EM) bridges the gap between the object sizes studied by X-ray
crystallography and light microscopy. 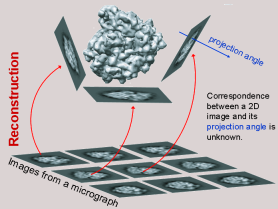 Single-particle EM is routinely
used to resolve the three-dimensional structure of large
macromolecular assemblies.
Single-particle EM is routinely
used to resolve the three-dimensional structure of large
macromolecular assemblies.
One of the major challenges in single-particle EM is structural
heterogeneity of the studied particles. Particles can adopt different
conformations and can be found in assemblies with alternative
quaternary-structure. Computational methods for image processing and
three-dimensional structure determination play a crucial role in
single-particle EM. Most commonly used computational approaches assume
that the imaged particles have homogeneous shape and
quaternary-structure. When this assumption is violated, the product of
the three-dimensional reconstruction is one low resolution
structure. My aim is to improve and redesign various computational
stages of particle reconstruction, taking into account the
heterogeneity of the data.
Reconstruction of Multiple Structural Conformations of Macromolecular Complexes
We devised a new computational method that reveals the existence of
different conformational states from EM data of macromolecular
complexes.
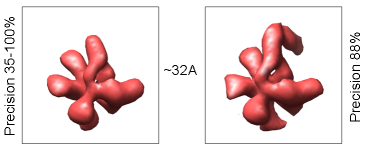 It is able to automatically classify the experimental
images into homogeneous subsets which produce structurally different
models. The method achieves high accuracy on synthetic data sets and
real data. In one test case, we
successfully differentiated between the real experimental images of
human translation initiation factor eIF3 and of eIF3 complexed with
hepatitis C virus (HCV) IRES RNA. In another experiment, where we used
experimental images of human RNA polymerase II, our method produced
two models that show a substantial conformational flexibility of the
protein complex.
It is able to automatically classify the experimental
images into homogeneous subsets which produce structurally different
models. The method achieves high accuracy on synthetic data sets and
real data. In one test case, we
successfully differentiated between the real experimental images of
human translation initiation factor eIF3 and of eIF3 complexed with
hepatitis C virus (HCV) IRES RNA. In another experiment, where we used
experimental images of human RNA polymerase II, our method produced
two models that show a substantial conformational flexibility of the
protein complex.
A Method for the Alignment of Heterogeneous Macromolecules
We proposed a feature-based image alignment method (Shatsky et al. '09) for single-particle
EM that is able to accommodate various similarity
scoring functions while efficiently sampling the two-dimensional
transformational space.
 We use this image alignment method to evaluate
the performance of a scoring function that is based on the Mutual
Information (MI) of two images rather than one that is based on the
commonly used cross-correlation function.
We show that alignment using MI for the
scoring function has far less model-dependent bias than is found with
cross-correlation based alignment. We also demonstrate that MI
improves the alignment of some types of heterogeneous data, provided
that the signal-to-noise ratio is relatively high. These results
indicate, therefore, that use of MI as the scoring function is well
suited for the alignment of class-averages computed from
single-particle images. Our method is tested on data from three model
structures and one real dataset.
We use this image alignment method to evaluate
the performance of a scoring function that is based on the Mutual
Information (MI) of two images rather than one that is based on the
commonly used cross-correlation function.
We show that alignment using MI for the
scoring function has far less model-dependent bias than is found with
cross-correlation based alignment. We also demonstrate that MI
improves the alignment of some types of heterogeneous data, provided
that the signal-to-noise ratio is relatively high. These results
indicate, therefore, that use of MI as the scoring function is well
suited for the alignment of class-averages computed from
single-particle images. Our method is tested on data from three model
structures and one real dataset.
Software:
Contributed to:
Publications:
D. Zhi, M. Shatsky, SE Brenner. 2010. Alignment-free local structural search by writhe decomposition. Bioinformatics. in press.
M. Shatsky, R.J. Hall, E. Nogales, J. Malik, S.E. Brenner. Automated multi-model reconstruction from single-particle electron microscopy data. J Struct Bio. 170:98-108, 2010.
BG Han, M. Dong, H. Liu, L. Camp, J. Geller, M. Singer, TC Hazen, M. Choi, HE Witkowsa, DA Ball, D. Typke, KH Downing, M. Shatsky, SE Brenner, JM Chandonia, MD Biggin, RM Glaeser. Survey of large protein complexes in D. vulgaris reveals unexpected structural diversity. Proceedings of the National Academy of Sciences of the United States of America 106:16580-16585. 2009.
M. Shatsky, R.J. Hall, S.E. Brenner, and R.M. Glaeser. A method for the alignment of heterogeneous macromolecules from electron microscopy. J Struct Bio. 166:67-78, 2009.
A. Shulman-Peleg, M.Shatsky, R.Nussinov and H.J.Wolfson. Prediction of Interacting Single-Stranded RNA Bases by Protein-Binding Patterns. Journal of Molecular Biology, 379(2):299-316, 2008.
A. Shulman-Peleg, M.Shatsky, R.Nussinov and H.J.Wolfson. Spatial chemical conservation of hot spot interactions in protein-protein complexes, BMC Biology, 5(1):43, 2007
D. Zhi, M. Shatsky, S. Brenner. Alignment-Free Local Structural Search by Writhe Decomposition. (abstract) Workshop on Algorithms in Bioinformatics, Springer Verlag, Lecture Notes in Computer Science, 2007.
K. Lasker, O. Dror, M.Shatsky, R.Nussinov, H.Wolfson. EMatch: Discovery of High Resolution Structural Homologues of Protein Domains In Intermediate Resolution Cryo-EM. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 4(1):28-39, 2007.
M. Shatsky, A. Shulman-Peleg, R. Nussinov, H. Wolfson. The Multiple Common Point Set Problem and its Application to Molecule Binding Pattern Detection. Journal of Computational Biology, 13(2):407-28, 2006.
M. Shatsky, R. Nussinov, and H.J. Wolfson. Optimization of Multiple Sequence Alignment Based on Multiple Structure Alignment. Proteins: Structure, Function, and Bioinformatics, 62(1):209-17, 2006.
A. Shulman-Peleg, M. Shatsky, R. Nussinov, H. Wolfson. MAPPIS: Multiple 3D Alignment of Protein-Protein Interfaces. M.R. Berthold et al. (Eds.): CompLife 2005, Lecture Notes in Computer Science, Volume 3695, pp. 91-103, 2005.
M. Shatsky, A. Shulman-Peleg, R. Nussinov, H. Wolfson. Recognition of Binding Patterns Common to a Set of Protein Structures. RECOMB 2005, Lecture Notes in Computer Science, Springer, vol. 3500, pp. 440-455, 2005.
M. Shatsky, R. Nussinov, H. Wolfson. A Method for Simultaneous Alignment of Multiple Protein Structures. Proteins: Structure, Function, and Genetics, 56(1), 143-156, 2004.
M. Shatsky, R. Nussinov, H. Wolfson. FlexProt: Alignment of Flexible Protein Structures Without a Pre-definition of Hinge Regions. Journal of Computational Biology, 11(1), 83-106, 2004.
D. Schneidman-Duhovny, Y. Inbar, V. Polak, M. Shatsky, I. Halperin, H. Benyamini, A. Barzilay, O. Shem-Tov, N. Haspel, R. Nussinov, H. J. Wolfson. Taking Geometry to its Edge: Fast Unbound Rigid (and Hinge-bent) Docking. Proteins: Structure, Function, and Genetics, 52(1), 107-112, 2003.
M. Shatsky, H.J. Wolfson, R.Nussinov. MultiProt - a Multiple Protein Structural Alignment Algorithm. Workshop on Algorithms in Bioinformatics, Springer Verlag, Lecture Notes in Computer Science 2452: 235-250, 2002.
M. Shatsky, R. Nussinov, H. Wolfson. Flexible protein alignment and hinge detection. Proteins: Structure, Function, and Genetics,48:242-256, 2002.
B. Ma, M. Shatsky, H.J. Wolfson, R.Nussinov. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Sci., 11, 184-197, 2002.
M. Shatsky, Z. Fligelman, R. Nussinov, H.J. Wolfson. Alignment of Flexible Protein Structures. Proc. 8th International Conference on Intelligent Systems for Molecular Biology (ISMB'00), 329-343, The AAAI press, 2000.
D. Zhi, M. Shatsky, SE Brenner. 2010. Alignment-free local structural search by writhe decomposition. Bioinformatics. in press.
M. Shatsky, R.J. Hall, E. Nogales, J. Malik, S.E. Brenner. Automated multi-model reconstruction from single-particle electron microscopy data. J Struct Bio. 170:98-108, 2010.
BG Han, M. Dong, H. Liu, L. Camp, J. Geller, M. Singer, TC Hazen, M. Choi, HE Witkowsa, DA Ball, D. Typke, KH Downing, M. Shatsky, SE Brenner, JM Chandonia, MD Biggin, RM Glaeser. Survey of large protein complexes in D. vulgaris reveals unexpected structural diversity. Proceedings of the National Academy of Sciences of the United States of America 106:16580-16585. 2009.
M. Shatsky, R.J. Hall, S.E. Brenner, and R.M. Glaeser. A method for the alignment of heterogeneous macromolecules from electron microscopy. J Struct Bio. 166:67-78, 2009.
A. Shulman-Peleg, M.Shatsky, R.Nussinov and H.J.Wolfson. Prediction of Interacting Single-Stranded RNA Bases by Protein-Binding Patterns. Journal of Molecular Biology, 379(2):299-316, 2008.
A. Shulman-Peleg, M.Shatsky, R.Nussinov and H.J.Wolfson. Spatial chemical conservation of hot spot interactions in protein-protein complexes, BMC Biology, 5(1):43, 2007
D. Zhi, M. Shatsky, S. Brenner. Alignment-Free Local Structural Search by Writhe Decomposition. (abstract) Workshop on Algorithms in Bioinformatics, Springer Verlag, Lecture Notes in Computer Science, 2007.
K. Lasker, O. Dror, M.Shatsky, R.Nussinov, H.Wolfson. EMatch: Discovery of High Resolution Structural Homologues of Protein Domains In Intermediate Resolution Cryo-EM. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 4(1):28-39, 2007.
M. Shatsky, A. Shulman-Peleg, R. Nussinov, H. Wolfson. The Multiple Common Point Set Problem and its Application to Molecule Binding Pattern Detection. Journal of Computational Biology, 13(2):407-28, 2006.
M. Shatsky, R. Nussinov, and H.J. Wolfson. Optimization of Multiple Sequence Alignment Based on Multiple Structure Alignment. Proteins: Structure, Function, and Bioinformatics, 62(1):209-17, 2006.
A. Shulman-Peleg, M. Shatsky, R. Nussinov, H. Wolfson. MAPPIS: Multiple 3D Alignment of Protein-Protein Interfaces. M.R. Berthold et al. (Eds.): CompLife 2005, Lecture Notes in Computer Science, Volume 3695, pp. 91-103, 2005.
M. Shatsky, A. Shulman-Peleg, R. Nussinov, H. Wolfson. Recognition of Binding Patterns Common to a Set of Protein Structures. RECOMB 2005, Lecture Notes in Computer Science, Springer, vol. 3500, pp. 440-455, 2005.
M. Shatsky, R. Nussinov, H. Wolfson. A Method for Simultaneous Alignment of Multiple Protein Structures. Proteins: Structure, Function, and Genetics, 56(1), 143-156, 2004.
M. Shatsky, R. Nussinov, H. Wolfson. FlexProt: Alignment of Flexible Protein Structures Without a Pre-definition of Hinge Regions. Journal of Computational Biology, 11(1), 83-106, 2004.
D. Schneidman-Duhovny, Y. Inbar, V. Polak, M. Shatsky, I. Halperin, H. Benyamini, A. Barzilay, O. Shem-Tov, N. Haspel, R. Nussinov, H. J. Wolfson. Taking Geometry to its Edge: Fast Unbound Rigid (and Hinge-bent) Docking. Proteins: Structure, Function, and Genetics, 52(1), 107-112, 2003.
M. Shatsky, H.J. Wolfson, R.Nussinov. MultiProt - a Multiple Protein Structural Alignment Algorithm. Workshop on Algorithms in Bioinformatics, Springer Verlag, Lecture Notes in Computer Science 2452: 235-250, 2002.
M. Shatsky, R. Nussinov, H. Wolfson. Flexible protein alignment and hinge detection. Proteins: Structure, Function, and Genetics,48:242-256, 2002.
B. Ma, M. Shatsky, H.J. Wolfson, R.Nussinov. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Sci., 11, 184-197, 2002.
M. Shatsky, Z. Fligelman, R. Nussinov, H.J. Wolfson. Alignment of Flexible Protein Structures. Proc. 8th International Conference on Intelligent Systems for Molecular Biology (ISMB'00), 329-343, The AAAI press, 2000.
Book Chapters and Reviews:
M. Shatsky, R. Nussinov, and H.J. Wolfson. Algorithms for Multiple Protein Structure Alignment and Structure-Derived Multiple Sequence Alignment. Protein Structure Prediction Series: Methods in Molecular Biology , Vol. 413. Zaki, Mohammed; Bystroff, Chris (Eds.), 2nd ed., 2007. Springer, Humana Press.
H. Wolfson, M. Shatsky, D. Duhovny, O. Dror, A. Shulman, B.Ma, R. Nussinov. From structure to function: methods and applications. Curr Protein Pept Sci., 6(2):171-83, 2005.
Software/Web-servers:
A. Shulman-Peleg, M.Shatsky, R.Nussinov and H.J.Wolfson. MultiBind and MAPPIS: webservers for multiple alignment of protein 3D-binding sites and their interactions. Nucleic Acids Res. 36(Web Server issue):W260-4, 2008.
M. Shatsky, O. Dror, D. Duhovny, A. Shulman, R. Nussinov, H. Wolfson. BioInfo3D: A Suite of Tools for Structural Bioinformatics. Nucleic Acid Research 32, 503-507, 2004.