| [Brenner Computational Biology Research Group] [Ed Green] |
Print version of this site [PDF 2.7 Mb]
|
This page is intended to serve as background and complement for the manuscript:
Lewis BP, Green RE, Brenner SE. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proceedings of the National Academy of Sciences of the United States of America 100:189-192. [PDF 0.25 Mb].
Alternative splicing has been shown to affect more than one-third of all human genes. We have found that many alternative isoforms are apparent targets of nonsense-mediated mRNA decay (NMD), an mRNA surveillance system. The coupling of alternative splicing with NMD is intriguing and could provide a general means of regulating gene expression.
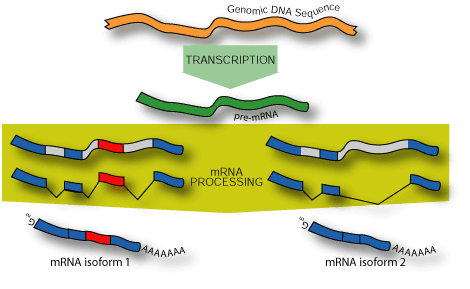
Differences between isoforms of an alternatively spliced gene may be
subtle or profound. For example, the human Bcl-x gene can be processed
to generate several isoforms with markedly different activities.
The Bcl-x(L) isoform inhibits apoptosis, whereas Bcl-x(S) can induce apoptosis[10].
Many other genes are alternatively spliced to produce isoforms whose differences
are only in non-coding regions; indeed, a recent study of alternative isoforms
in mice showed that 21% of splice variations do not affect coding potential[14,
15].
Because of the prevalence of alternative splicing, researchers would like
to know the regulatory mechanisms that control it and the functional consequences
of the isoforms that are produced. To these ends, several groups
have classified and catagorized known alternative isoforms in terms of
changes in gene structure between alternative isoforms or by the functional
classes of the genes that are involved[4,
16].
The only general conclusion that can be drawn from these analyses, however,
is that alternative splicing affects genes of nearly every functional class
by modifying gene structure in every conceivable way, such as using mutually
exclusive exons or alternative donor sites.
After mRNA processing, most transcripts are exported to the cytoplasm
for translation into protein. Each mRNA transcript can serve as template
for repeated translation into protein by ribosomes. The number of protein
products produced by any single mRNA can vary widely. This number is a
function of, among other things, the life span of the mRNA. In the cytoplasm,
mRNAs gradually loose their poly-adenosine tails. Once this tail has been
reduced to a threshold length, the mRNA is digested by exonucleases. Specific
signal sequences, AREs for example[17], can affect the
rate at which the poly-adenosine tail is shortened. Some mRNAs, however,
can be degraded almost immediately, by a process that is independent of
poly-adenosine tail length.

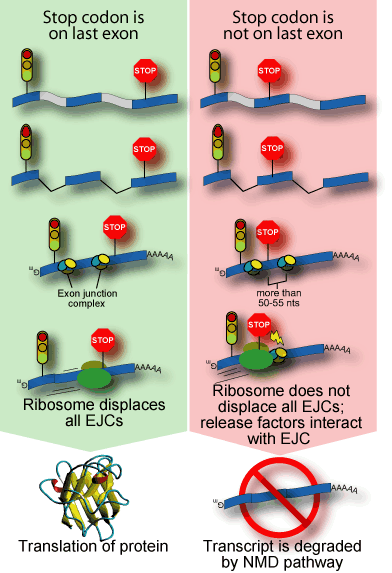
It has long been known that mRNAs carrying a premature termination codon are highly unstable[18-24]. A process known as nonsense-mediated mRNA decay (NMD) recognizes these mRNAs and degrades them. Recently, the molecular details of this process have begun to be elucidated. During mRNA processing, a complex is deposited near sites of intron removal[25-30]. These exon-junction complexes are important both for facilitating export from the nucleus and for remembering gene structure[31]. That is, they mark the sites where the introns were spliced out. This relative positioning appears to be checked during the pioneering round of translation[32, 33]. The ribosome, as it traverses the mRNA, displaces any exon-junction complexes in its path. Upon arrival at the termination codon, release factors interact with any undisplaced exon-junction complexes[34]. This association triggers decapping of the transcript, followed by degradation[35].
In vertebrates, the location of the last exon-junction complex relative to the termination codon usually determines whether the transcript will be subjected to NMD or not. If the termination codon is downstream of or within about 50 nucleotides of the final exon-junction complex then the transcript is translated normally. However, if the termination codon is further than about 50 nucleotides upstream of any exon-junction complexes, then the transcript is down regulated by NMD.
There are several lines of evidence supporting this model. First, intron-less
transcripts appear to be generally immune to NMD[36-38].
Second, tethering any of several components of the exon-junction complex
downstream of a termination codon will cause the transcript to be degraded[35].  Finally, NMD is inhibited by cis-elements or chemical reagents that
prevent efficient translation[23,
39].
Finally, NMD is inhibited by cis-elements or chemical reagents that
prevent efficient translation[23,
39].
This model of NMD has led to increased understanding of the formerly mystifying relationship between genotype and phenotype for many disease genes like dystrophin[40] and beta-globin[41, 42].
Analysis of the well characterized human genes in RefSeq reveals that
the vast majority are not candidates for NMD[16,
49]
. This is because their termination codons are on the last exon or within
50 nucleotides of it. This indicates that NMD is pervasive, as there appears
to be selective pressure toward keeping the termination codon on the final
exon. Start codons, on the other hand are commonly found downstream of
the first intron.
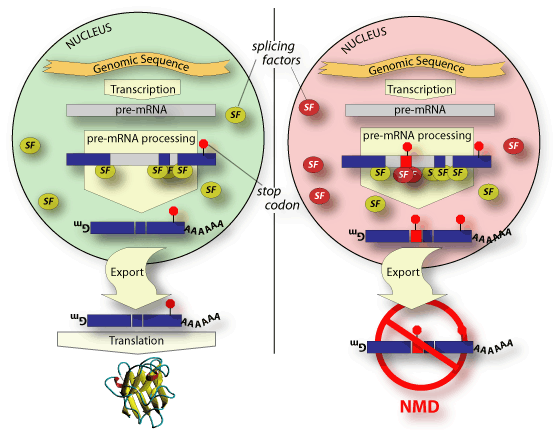
Alternatively spliced genes may have some isoforms that are candidates
for NMD and others that are translated normally. By coupling alternative
splicing to NMD, a cell could functionally down regulate expression of
that gene under desired conditions. In these cases, the protein coding
sequence of the alternative isoform is not nearly as important as the fact
that its structure will cause it to be degraded by NMD. There are,
in fact, cases in which alternative splicing does not affect the coding
region at all. It only affects whether the isoform will be down regulated
by NMD.
Regulation of this kind, which we term regulated unproductive splicing and translation (RUST), is mediated by the splice environment - the set of splicing factors present and active at a given time and place. Under certain conditions, one set of splice sites could be used that generate an isoform whose stop codon is on the last exon. This productive isoform would then be translated normally. Under different conditions or in a different cell, alternative splice sites could be used that introduce a premature termination codon, generating an unproductive isoform. This can be done by splicing in an alternative exon (as in the figure), causing a frameshift, or splicing out an intron downstream of the normal termination codon. This would shunt the gene from the normally translated pathway into the NMD pathway.
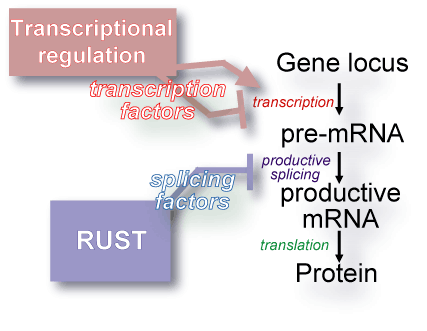
RUST is analogous to transcriptional regulation in that both cis-elements
and trans-factors are involved. Under transcriptional regulation,
transcription factors interact with the
cis-control elements in
the regulatory regions of target genes. The concentration, localization,
and activity of transcription factors determine which genes will be transcribed
into pre-mRNA. RUST acts during pre-mRNA processing, the next step in gene
expression. As with transcriptional regulation, the concentration,
localization, and activity of trans-factors determines which genes
will generate functional end products. In this case, however, the
trans-factors
are splicing factors and the
cis-elements are the splicing signals
present within the pre-mRNAs. Several well characterized signaling
pathways have been shown to alter the splice environment by activating
splicing factors[50,51]. Furthermore,
the cis-elements needed for RUST are well conserved in several known
RUST genes [52,
53]. In some
cases, these are even more conserved than the protein coding sequence.

Recent studies by several independent research groups have uncovered
genes whose expression appears to be influenced by RUST [13,
46,
47,
48].
One particularly interesting example is the splicing factor, SC35
[11-13], which autoregulates its own
expression by coupling alternative splicing with NMD.
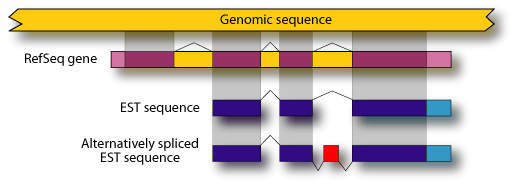
Although some alternative isoforms are described in RefSeq[43] and other databases, the majority are not. The most comprehensive data sources for alternative splicing are the EST databases, such as dbEST[44]. Several groups have shown that it is possible to cluster EST sequences with one another or with known gene sequence to learn which transcripts are alternatively spliced and what these alternative isoforms look like. With human genome sequence available, it is then possible to compare these alternative isoforms with their genomic regions to determine their underlying gene structures. This information can then be used to predict which isoforms of a given gene do not follow the 50 nucleotide rule and are therefore candidates for NMD.
To determine the extent to which alternative splicing generates
NMD-candidate isoforms, we aligned RefSeq sequences to the human genome[45]
to determine their gene structures. To the coding region of these alignments,
we then aligned EST sequences to reveal patterns of alternative splicing.
If the EST sequences showed a different splicing pattern than the RefSeq
sequence, it was taken as evidence for an alternatively spliced isoform. 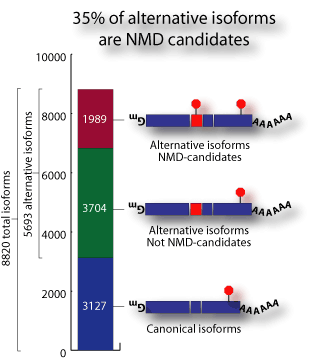 Many
filters were applied to ensure reliability. For example, we disregarded
cases of intron retention as these are indistinguishable from incompletely
processed transcripts, a common EST database contaminant. We also restricted
alignments to the coding regions of the RefSeq sequences to ensure alignments
of the highest quality possible. Because the RefSeq isoforms are annotated
with start and stop codon positions, it was then possible to determine
which isoforms obeyed the 50 nucleotide NMD rule.
Many
filters were applied to ensure reliability. For example, we disregarded
cases of intron retention as these are indistinguishable from incompletely
processed transcripts, a common EST database contaminant. We also restricted
alignments to the coding regions of the RefSeq sequences to ensure alignments
of the highest quality possible. Because the RefSeq isoforms are annotated
with start and stop codon positions, it was then possible to determine
which isoforms obeyed the 50 nucleotide NMD rule.
We found that about one third of all alternative splicing events generate
NMD candidate isoforms. Furthermore, about one third of all genes for which
there is alternative splicing EST data generate at least one NMD candidate
isoform. These numbers suggest that coupling of alternative splicing and
NMD may be widespread. Because our analysis did not consider alternative
splicing outside of coding regions and because destabilized transcripts
may be under-represented in EST databases, it could be the case that this
phenomenon is even more pervasive than our data suggest.
Another possible conclusion is that the process of splicing is not nearly as precise as one might imagine. Perhaps the process of finding and splicing small exons in a sea of large introns is so difficult that the splicing machinery is very error prone. If this is the case, then the splicing process may rely on the presence of the NMD pathway to dispose of incompletely or incorrectly spliced products to an extent not previously appreciated. We cannot presently rule out this possibility. Therefore, it is imperative that researchers who use the EST databases as a source of gene sequence must consider which isoforms are NMD candidates. We feel that this is especially prudent advice, as genes are commonly cloned as intronless cDNAs, immune to NMD, prior to further characterization.
It is also possible, and likely, that there are still gaps in our understanding of the NMD pathway. There are a handful of genes that generate isoforms that should be NMD substrate, based on the 50 nucleotide rule, that appear to be immune to NMD (the male-specific isoform of sex-lethal in drosophila, for example). Also, there are specific signal sequences that appear to be functionally equivalent to exon-junction complexes in triggering NMD. It is likely that there are caveats to the 50 nucleotide rule that, once discovered, can be used to refine our list of NMD-candidate isoforms.
A final, intriguing possibility is that the regulated coupling of alternative splicing and NMD represents a general mode of controlling gene expression. This interpretation is attractive in that it depends only on systems, NMD and alternative splicing, that are known to be pervasive. In the RUST process, splicing factors play a role analogous to transcription factors in that they regulate which genes are expressed. In addition to being attractive just for its ease of use, RUST would allow for a degree of temporal control of very large genes that take a long time to transcribe, that is unachievable with transcription factors. Several instances of RUST have already been discovered, like the splicing factors SC35[13] and AUF1[46].
J. S. Anderson and R. Parker, "RNA turnover: the helicase story
unwinds," Curr Biol, vol. 6, pp. 780-2, 1996.
S. E. Applequist, M. Selg, C. Raman, and H. M. Jack, "Cloning and characterization
of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing
UPF1 protein," Nucleic Acids Res, vol. 25, pp. 814--21, 1997.
L. Aravind and E. V. Koonin, "Eukaryote-specific domains in translation
initiation factors: implications for translation regulation and evolution
of the translation system," Genome Res, vol. 10, pp. 1172-84, 2000.
R. Aronoff, R. Baran, and J. Hodgkin, "Molecular identification of
smg-4, required for mRNA surveillance in C. elegans," Gene, vol. 268, pp.
153-64, 2001.
R. Asselta, S. Duga, S. Spena, E. Santagostino, F. Peyvandi, G. Piseddu,
R. Targhetta, M. Malcovati, P. M. Mannucci, and M. L. Tenchini, "Congenital
afibrinogenemia: mutations leading to premature termination codons in fibrinogen
A alpha-chain gene are not associated with the decay of the mutant mRNAs,"
Blood, vol. 98, pp. 3685-92, 2001.
A. L. Atkin, N. Altamura, P. Leeds, and M. R. Culbertson, "The majority
of yeast UPF1 co-localizes with polyribosomes in the cytoplasm," Mol Biol
Cell, vol. 6, pp. 611-25, 1995.
A. L. Atkin, L. R. Schenkman, M. Eastham, J. N. Dahlseid, M. J. Lelivelt,
and M. R. Culbertson, "Relationship between yeast polyribosomes and Upf
proteins required for nonsense mRNA decay," J Biol Chem, vol. 272, pp.
22163-72, 1997.
C. A. Barnes, "Upf1 and Upf2 proteins mediate normal yeast mRNA degradation
when translation initiation is limited," Nucleic Acids Res, vol. 26, pp.
2433-41, 1998.
E. R. Barton-Davis, L. Cordier, D. I. Shoturma, S. E. Leland, and H.
L. Sweeney, "Aminoglycoside antibiotics restore dystrophin function to
skeletal muscles of mdx mice," J Clin Invest, vol. 104, pp. 375-81, 1999.
J. F. Bateman, S. Freddi, S. R. Lamande, P. Byers, S. Nasioulas, J.
Douglas, R. Otway, M. Kohonen-Corish, E. Edkins, and S. Forrest, "Reliable
and sensitive detection of premature termination mutations using a protein
truncation test designed to overcome problems of nonsense-mediated mRNA
instability," Hum Mutat, vol. 13, pp. 311-7, 1999.
P. Belgrader, J. Cheng, and L. E. Maquat, "Evidence to implicate translation
by ribosomes in the mechanism by which nonsense codons reduce the nuclear
level of human triosephosphate isomerase mRNA," Proc Natl Acad Sci U S
A, vol. 90, pp. 482-6, 1993.
P. Belgrader, J. Cheng, X. Zhou, L. S. Stephenson, and L. E. Maquat,
"Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life,"
Mol Cell Biol, vol. 14, pp. 8219-28, 1994.
P. Belgrader and L. E. Maquat, "Nonsense but not missense mutations
can decrease the abundance of nuclear mRNA for the mouse major urinary
protein, while both types of mutations can facilitate exon skipping," Mol
Cell Biol, vol. 14, pp. 6326-36, 1994.
J. P. Belk, F. He, and A. Jacobson, "Overexpression of truncated Nmd3p
inhibits protein synthesis in yeast," Rna, vol. 5, pp. 1055-70, 1999.
A. Bhattacharya, K. Czaplinski, P. Trifillis, F. He, A. Jacobson, and
S. W. Peltz, "Characterization of the biochemical properties of the human
Upf1 gene product that is involved in nonsense-mediated mRNA decay," Rna,
vol. 6, pp. 1226-35, 2000.
L. Bidou, G. Stahl, I. Hatin, O. Namy, J. P. Rousset, and P. J. Farabaugh,
"Nonsense-mediated decay mutants do not affect programmed-1 frameshifting,"
Rna-a Publication of the Rna Society, vol. 6, pp. 952-961, 2000.
A. T. Bond, D. A. Mangus, F. He, and A. Jacobson, "Absence of Dbp2p
alters both nonsense-mediated mRNA decay and rRNA processing," Mol Cell
Biol, vol. 21, pp. 7366-79, 2001.
K. S. Brocke, G. Neu-Yilik, N. H. Gehring, M. W. Hentze, and A. E.
Kulozik, "The human intronless melanocortin 4-receptor gene is NMD insensitive,"
Hum Mol Genet, vol. 11, pp. 331-5, 2002.
M. Buhler, M. F. Wilkinson, and O. Muhlemann, "Intranuclear degradation
of nonsense codon-containing mRNA," EMBO Rep, vol. 3, pp. 646-51, 2002.
P. H. Byers, "Killing the messenger: new insights into nonsense-mediated
mRNA decay," J Clin Invest, vol. 109, pp. 3-6, 2002.
B. M. Cali and P. Anderson, "mRNA surveillance mitigates genetic dominance
in Caenorhabditis elegans," Mol Gen Genet, vol. 260, pp. 176-84, 1998.
B. M. Cali, S. L. Kuchma, J. Latham, and P. Anderson, "smg-7 is required
for mRNA surveillance in Caenorhabditis elegans," Genetics, vol. 151, pp.
605-16, 1999.
L. Cartegni, S. L. Chew, and A. R. Krainer, "Listening to silence and
understanding nonsense: exonic mutations that affect splicing," Nat Rev
Genet, vol. 3, pp. 285-98, 2002.
M. S. Carter, J. Doskow, P. Morris, S. Li, R. P. Nhim, S. Sandstedt,
and M. F. Wilkinson, "A regulatory mechanism that detects premature nonsense
codons in T-cell receptor transcripts in vivo is reversed by protein synthesis
inhibitors in vitro," J Biol Chem, vol. 270, pp. 28995-9003, 1995.
M. S. Carter, S. Li, and M. F. Wilkinson, "A splicing-dependent regulatory
mechanism that detects translation signals," Embo J, vol. 15, pp. 5965-75,
1996.
J. Cheng, P. Belgrader, X. Zhou, and L. E. Maquat, "Introns are cis
effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance,"
Mol Cell Biol, vol. 14, pp. 6317-25, 1994.
J. Cheng, M. Fogel-Petrovic, and L. E. Maquat, "Translation to near
the distal end of the penultimate exon is required for normal levels of
spliced triosephosphate isomerase mRNA," Mol Cell Biol, vol. 10, pp. 5215-25,
1990.
J. Cheng and L. E. Maquat, "Nonsense codons can reduce the abundance
of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life
of cytoplasmic mRNA," Mol Cell Biol, vol. 13, pp. 1892-902, 1993.
P. M. Clissold and C. P. Ponting, "PIN domains in nonsense-mediated
mRNA decay and RNAi," Curr Biol, vol. 10, pp. R888-90, 2000.
Y. Cui, J. D. Dinman, and S. W. Peltz, "Mof4-1 is an allele of the
UPF1/IFS2 gene which affects both mRNA turnover and -1 ribosomal frameshifting
efficiency," Embo J, vol. 15, pp. 5726-36, 1996.
Y. Cui, K. W. Hagan, S. Zhang, and S. W. Peltz, "Identification and
characterization of genes that are required for the accelerated degradation
of mRNAs containing a premature translational termination codon," Genes
Dev, vol. 9, pp. 423-36, 1995.
M. R. Culbertson, "RNA surveillance. Unforeseen consequences for gene
expression, inherited genetic disorders and cancer," Trends Genet, vol.
15, pp. 74-80, 1999.
M. R. Culbertson, L. Charnas, M. T. Johnson, and G. R. Fink, "Frameshifts
and frameshift suppressors in Saccharomyces cerevisiae," Genetics, vol.
86, pp. 745-64, 1977.
M. R. Culbertson, K. M. Underbrink, and G. R. Fink, "Frameshift suppression
Saccharomyces cerevisiae. II. Genetic properties of group II suppressors,"
Genetics, vol. 95, pp. 833-53, 1980.
K. Czaplinski, N. Majlesi, T. Banerjee, and S. W. Peltz, "Mtt1 is a
Upf1-like helicase that interacts with the translation termination factors
and whose overexpression can modulate termination efficiency," Rna, vol.
6, pp. 730-43, 2000.
K. Czaplinski, M. J. Ruiz-Echevarria, C. I. Gonzalez, and S. W. Peltz,
"Should we kill the messenger? The role of the surveillance complex in
translation termination and mRNA turnover," Bioessays, vol. 21, pp. 685-96,
1999.
K. Czaplinski, M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng,
H. A. Perlick, H. C. Dietz, M. D. Ter-Avanesyan, and S. W. Peltz, "The
surveillance complex interacts with the translation release factors to
enhance termination and degrade aberrant mRNAs," Genes Dev, vol. 12, pp.
1665-77, 1998.
K. Czaplinski, Y. Weng, K. W. Hagan, and S. W. Peltz, "Purification
and characterization of the Upf1 protein: a factor involved in translation
and mRNA degradation," Rna, vol. 1, pp. 610-23, 1995.
I. O. Daar and L. E. Maquat, "Premature translation termination mediates
triosephosphate isomerase mRNA degradation," Mol Cell Biol, vol. 8, pp.
802-13, 1988.
J. N. Dahlseid, J. Puziss, R. L. Shirley, A. L. Atkin, P. Hieter, and
M. R. Culbertson, "Accumulation of mRNA coding for the ctf13p kinetochore
subunit of Saccharomyces cerevisiae depends on the same factors that promote
rapid decay of nonsense mRNAs," Genetics, vol. 150, pp. 1019-35, 1998.
S. Danckwardt, G. Neu-Yilik, R. Thermann, U. Frede, M. W. Hentze, and
A. E. Kulozik, "Abnormally spliced beta-globin mRNAs: a single point mutation
generates transcripts sensitive and insensitive to nonsense-mediated mRNA
decay," Blood, vol. 99, pp. 1811-6, 2002.
B. Das, Z. Guo, P. Russo, P. Chartrand, and F. Sherman, "The role of
nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation,"
Mol Cell Biol, vol. 20, pp. 2827-38, 2000.
G. Denning, L. Jamieson, L. E. Maquat, E. A. Thompson, and A. P. Fields,
"Cloning of a novel phosphatidylinositol kinase-related kinase: characterization
of the human SMG-1 RNA surveillance protein," J Biol Chem, vol. 276, pp.
22709-14, 2001.
H. C. Dietz, "Nonsense mutations and altered splice-site selection,"
Am J Hum Genet, vol. 60, pp. 729-30, 1997.
H. C. Dietz, D. Valle, C. A. Francomano, R. J. Kendzior, Jr., R. E.
Pyeritz, and G. R. Cutting, "The skipping of constitutive exons in vivo
induced by nonsense mutations," Science, vol. 259, pp. 680-3, 1993.
T. Dunckley and R. Parker, "The DCP2 protein is required for mRNA decapping
in Saccharomyces cerevisiae and contains a functional MutT motif," Embo
J, vol. 18, pp. 5411-22, 1999.
T. Dunckley and R. Parker, "Yeast mRNA decapping enzyme," Methods Enzymol,
vol. 342, pp. 226-33, 2001.
T. Dunckley, M. Tucker, and R. Parker, "Two related proteins, Edc1p
and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae," Genetics,
vol. 157, pp. 27-37, 2001.
S. Freddi, R. Savarirayan, and J. F. Bateman, "Molecular diagnosis
of Stickler syndrome: a COL2A1 stop codon mutation screening strategy that
is not compromised by mutant mRNA instability," Am J Med Genet, vol. 90,
pp. 398-406, 2000.
P. A. Frischmeyer and H. C. Dietz, "Nonsense-mediated mRNA decay in
health and disease," Hum Mol Genet, vol. 8, pp. 1893-900, 1999.
M. Gao, C. J. Wilusz, S. W. Peltz, and J. Wilusz, "A novel mRNA-decapping
activity in HeLa cytoplasmic extracts is regulated by AU-rich elements,"
Embo J, vol. 20, pp. 1134-43, 2001.
C. I. Gonzalez, A. Bhattacharya, W. Wang, and S. W. Peltz, "Nonsense-mediated
mRNA decay in Saccharomyces cerevisiae," Gene, vol. 274, pp. 15-25, 2001.
C. I. Gonzalez, M. J. Ruiz-Echevarria, S. Vasudevan, M. F. Henry, and
S. W. Peltz, "The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript
for nonsense-mediated mRNA decay," Mol Cell, vol. 5, pp. 489-99, 2000.
J. P. Gudikote and M. F. Wilkinson, "T-cell receptor sequences that
elicit strong down-regulation of premature termination codon-bearing transcripts,"
Embo J, vol. 21, pp. 125-34, 2002.
K. W. Hagan, M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz, "Characterization
of cis-acting sequences and decay intermediates involved in nonsense-mediated
mRNA turnover," Mol Cell Biol, vol. 15, pp. 809-23, 1995.
F. He, A. H. Brown, and A. Jacobson, "Interaction between Nmd2p and
Upf1p is required for activity but not for dominant-negative inhibition
of the nonsense-mediated mRNA decay pathway in yeast," Rna, vol. 2, pp.
153-70, 1996.
F. He, A. H. Brown, and A. Jacobson, "Upf1p, Nmd2p, and Upf3p are interacting
components of the yeast nonsense-mediated mRNA decay pathway," Mol Cell
Biol, vol. 17, pp. 1580-94, 1997.
F. He and A. Jacobson, "Identification of a novel component of the
nonsense-mediated mRNA decay pathway by use of an interacting protein screen,"
Genes Dev, vol. 9, pp. 437-54, 1995.
F. He and A. Jacobson, "Upf1p, Nmd2p, and Upf3p regulate the decapping
and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type
mRNAs," Mol Cell Biol, vol. 21, pp. 1515-30, 2001.
F. He, S. W. Peltz, J. L. Donahue, M. Rosbash, and A. Jacobson, "Stabilization
and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant,"
Proc Natl Acad Sci U S A, vol. 90, pp. 7034-8, 1993.
P. Hilleren and R. Parker, "Mechanisms of mRNA surveillance in eukaryotes,"
Annu Rev Genet, vol. 33, pp. 229-60, 1999.
P. Hilleren and R. Parker, "mRNA surveillance in eukaryotes: kinetic
proofreading of proper translation termination as assessed by mRNP domain
organization?," Rna, vol. 5, pp. 711-9, 1999.
A. Huber, C. Yee, T. N. Darling, and K. B. Yancey, "Comprehensive analysis
of gene expression profiles in keratinocytes from patients with generalized
atrophic benign epidermolysis bullosa," Exp Dermatol, vol. 11, pp. 75-81,
2002.
Y. Ishigaki, X. J. Li, G. Serin, and L. E. Maquat, "Evidence for a
pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay
in mammalian cells are bound by CBP80 and CBP20," Cell, vol. 106, pp. 607-617,
2001.
A. Jacobson and S. W. Peltz, "Interrelationships of the pathways of
mRNA decay and translation in eukaryotic cells," Annu Rev Biochem, vol.
65, pp. 693-739, 1996.
A. Jacobson and S. W. Peltz, "Tools for turnover: methods for analysis
of mRNA stability in eukaryotic cells," Methods, vol. 17, pp. 1-2, 1999.
D. Jeganathan, M. F. Fox, J. M. Young, J. R. Yates, J. P. Osborne,
and S. Povey, "Nonsense-mediated RNA decay in the TSC1 gene suggests a
useful tool pre- and post-positional cloning," Hum Genet, vol. 111, pp.
555-65, 2002.
R. B. Jones, F. Wang, Y. Luo, C. Yu, C. Jin, T. Suzuki, M. Kan, and
W. L. McKeehan, "The nonsense-mediated decay pathway and mutually exclusive
expression of alternatively spliced FGFR2IIIb and -IIIc mRNAs," J Biol
Chem, vol. 276, pp. 4158-67, 2001.
T. Karl, K. Onder, R. Kodzius, A. Pichova, H. Wimmer, A. Th r, H. Hundsberger,
M. Loffler, T. Klade, A. Beyer, M. Breitenbach, and L. Koller, "GRC5 and
NMD3 function in translational control of gene expression and interact
genetically," Curr Genet, vol. 34, pp. 419-29, 1999.
V. N. Kim, N. Kataoka, and G. Dreyfuss, "Role of the nonsense-mediated
decay factor hUpf3 in the splicing-dependent exon-exon junction complex,"
Science, vol. 293, pp. 1832-1836, 2001.
V. N. Kim, J. Yong, N. Kataoka, L. Abel, M. D. Diem, and G. Dreyfuss,
"The Y14 protein communicates to the cytoplasm the position of exon-exon
junctions," Embo Journal, vol. 20, pp. 2062-2068, 2001.
A. J. Kinniburgh, L. E. Maquat, T. Schedl, E. Rachmilewitz, and J.
Ross, "mRNA-deficient beta o-thalassemia results from a single nucleotide
deletion," Nucleic Acids Res, vol. 10, pp. 5421-7, 1982.
H. Le Hir, D. Gatfield, I. C. Braun, D. Forler, and E. Izaurralde,
"The protein Mago provides a link between splicing and mRNA localization,"
EMBO Rep, vol. 2, pp. 1119-24, 2001.
H. Le Hir, D. Gatfield, E. Izaurralde, and M. J. Moore, "The exon-exon
junction complex provides a binding platform for factors involved in mRNA
export and nonsense-mediated mRNA decay," Embo Journal, vol. 20, pp. 4987-4997,
2001.
H. Le Hir, E. Izaurralde, L. E. Maquat, and M. J. Moore, "The spliceosome
deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon
junctions," Embo J, vol. 19, pp. 6860-9, 2000.
H. Le Hir, M. J. Moore, and L. E. Maquat, "Pre-mRNA splicing alters
mRNP composition: evidence for stable association of proteins at exon-exon
junctions," Genes & Development, vol. 14, pp. 1098-1108, 2000.
B. S. Lee and M. R. Culbertson, "Identification of an additional gene
required for eukaryotic nonsense mRNA turnover," Proc Natl Acad Sci U S
A, vol. 92, pp. 10354-8, 1995.
P. Leeds, S. W. Peltz, A. Jacobson, and M. R. Culbertson, "The product
of the yeast UPF1 gene is required for rapid turnover of mRNAs containing
a premature translational termination codon," Genes Dev, vol. 5, pp. 2303-14,
1991.
P. Leeds, J. M. Wood, B. S. Lee, and M. R. Culbertson, "Gene products
that promote mRNA turnover in Saccharomyces cerevisiae," Mol Cell Biol,
vol. 12, pp. 2165-77, 1992.
F. Lejeune, Y. Ishigaki, X. Li, and L. E. Maquat, "The exon junction
complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian
cells: dynamics of mRNP remodeling," Embo J, vol. 21, pp. 3536-45, 2002.
M. J. Lelivelt and M. R. Culbertson, "Yeast Upf proteins required for
RNA surveillance affect global expression of the yeast transcriptome,"
Mol Cell Biol, vol. 19, pp. 6710-9, 1999.
J. E. Lew, S. Enomoto, and J. Berman, "Telomere length regulation and
telomeric chromatin require the nonsense-mediated mRNA decay pathway,"
Mol Cell Biol, vol. 18, pp. 6121-30, 1998.
B. P. Lewis, R. E. Green, and S. E. Brenner, "Evidence for the widespread
coupling of alternative splicing and nonsense-mediated mRNA decay in humans,"
Proc Natl Acad Sci U S A, 2002.
S. Li, D. Leonard, and M. F. Wilkinson, "T cell receptor (TCR) mini-gene
mRNA expression regulated by nonsense codons: a nuclear-associated translation-like
mechanism," J Exp Med, vol. 185, pp. 985-92, 1997.
S. Li and M. F. Wilkinson, "Nonsense surveillance in lymphocytes?,"
Immunity, vol. 8, pp. 135-41, 1998.
S. Lim, J. J. Mullins, C. M. Chen, K. W. Gross, and L. E. Maquat, "Novel
metabolism of several beta zero-thalassemic beta-globin mRNAs in the erythroid
tissues of transgenic mice," Embo J, vol. 8, pp. 2613-9, 1989.
S. K. Lim, C. D. Sigmund, K. W. Gross, and L. E. Maquat, "Nonsense
codons in human beta-globin mRNA result in the production of mRNA degradation
products," Mol Cell Biol, vol. 12, pp. 1149-61, 1992.
R. M. Long, D. J. Elliott, F. Stutz, M. Rosbash, and R. H. Singer,
"Spatial consequences of defective processing of specific yeast mRNAs revealed
by fluorescent in situ hybridization," Rna, vol. 1, pp. 1071-8, 1995.
J. Lykke-Andersen, "mRNA quality control: Marking the message for life
or death," Curr Biol, vol. 11, pp. R88-91, 2001.
J. Lykke-Andersen, "Identification of a human decapping complex associated
with hUpf proteins in nonsense-mediated decay," Mol Cell Biol, vol. 22,
pp. 8114-21, 2002.
J. Lykke-Andersen, M. D. Shu, and J. A. Steitz, "Human Upf proteins
target an mRNA for nonsense-mediated decay when bound downstream of a termination
codon," Cell, vol. 103, pp. 1121-1131, 2000.
J. Lykke-Andersen, M. D. Shu, and J. A. Steitz, "Communication of the
position of exon-exon junctions to the mRNA surveillance machinery by the
protein RNPS1," Science, vol. 293, pp. 1836-1839, 2001.
A. B. Maderazo, F. He, D. A. Mangus, and A. Jacobson, "Upf1p control
of nonsense mRNA translation is regulated by Nmd2p and Upf3p," Mol Cell
Biol, vol. 20, pp. 4591-603, 2000.
S. E. Mango, "Stop making nonSense: the C. elegans smg genes," Trends
Genet, vol. 17, pp. 646-53, 2001.
D. A. Mangus, N. Amrani, and A. Jacobson, "Pbp1p, a factor interacting
with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation,"
Mol Cell Biol, vol. 18, pp. 7383-96, 1998.
D. A. Mangus and A. Jacobson, "Linking mRNA turnover and translation:
assessing the polyribosomal association of mRNA decay factors and degradative
intermediates," Methods, vol. 17, pp. 28-37, 1999.
L. E. Maquat, "When cells stop making sense: effects of nonsense codons
on RNA metabolism in vertebrate cells," Rna, vol. 1, pp. 453-65, 1995.
L. E. Maquat, "Defects in RNA splicing and the consequence of shortened
translational reading frames," Am J Hum Genet, vol. 59, pp. 279-86, 1996.
L. E. Maquat, "The power of point mutations," Nat Genet, vol. 27, pp.
5-6, 2001.
L. E. Maquat, "Evidence that selenium deficiency results in the cytoplasmic
decay of GPx1 mRNA dependent on pre-mRNA splicing proteins bound to the
mRNA exon-exon junction," Biofactors, vol. 14, pp. 37-42, 2001.
L. E. Maquat, "Nonsense-mediated mRNA decay," Curr Biol, vol. 12, pp.
R196-7, 2002.
L. E. Maquat and G. G. Carmichael, "Quality control of mRNA function,"
Cell, vol. 104, pp. 173-6, 2001.
L. E. Maquat, R. Chilcote, and P. M. Ryan, "Human triosephosphate isomerase
cDNA and protein structure. Studies of triosephosphate isomerase deficiency
in man," J Biol Chem, vol. 260, pp. 3748-53, 1985.
L. E. Maquat and A. J. Kinniburgh, "A beta zero-thalassemic beta-globin
RNA that is labile in bone marrow cells is relatively stable in HeLa cells,"
Nucleic Acids Res, vol. 13, pp. 2855-67, 1985.
L. E. Maquat, A. J. Kinniburgh, L. R. Beach, G. R. Honig, J. Lazerson,
W. B. Ershler, and J. Ross, "Processing of human beta-globin mRNA precursor
to mRNA is defective in three patients with beta+-thalassemia," Proc Natl
Acad Sci U S A, vol. 77, pp. 4287-91, 1980.
L. E. Maquat, A. J. Kinniburgh, E. A. Rachmilewitz, and J. Ross, "Unstable
beta-globin mRNA in mRNA-deficient beta o thalassemia," Cell, vol. 27,
pp. 543-53, 1981.
L. E. Maquat and X. Li, "Mammalian heat shock p70 and histone H4 transcripts,
which derive from naturally intronless genes, are immune to nonsense-mediated
decay," Rna, vol. 7, pp. 445-56, 2001.
I. McIntosh, A. Hamosh, and H. C. Dietz, "Nonsense mutations and diminished
mRNA levels," Nat Genet, vol. 4, pp. 219, 1993.
S. M. Medghalchi, P. A. Frischmeyer, J. T. Mendell, A. G. Kelly, A.
M. Lawler, and H. C. Dietz, "Rent1, a trans-effector of nonsense-mediated
mRNA decay, is essential for mammalian embryonic viability," Hum Mol Genet,
vol. 10, pp. 99-105, 2001.
J. T. Mendell, C. M. ap Rhys, and H. C. Dietz, "Separable roles for
rent1/hUpf1 in altered splicing and decay of nonsense transcripts," Science,
vol. 298, pp. 419-22, 2002.
J. T. Mendell and H. C. Dietz, "When the message goes awry: disease-producing
mutations that influence mRNA content and performance," Cell, vol. 107,
pp. 411-4, 2001.
J. T. Mendell, S. M. Medghalchi, R. G. Lake, E. N. Noensie, and H.
C. Dietz, "Novel Upf2p orthologues suggest a functional link between translation
initiation and nonsense surveillance complexes," Mol Cell Biol, vol. 20,
pp. 8944-57, 2000.
P. Mitchell and D. Tollervey, "mRNA turnover," Curr Opin Cell Biol,
vol. 13, pp. 320-5, 2001.
Q. M. Mitrovich and P. Anderson, "Unproductively spliced ribosomal
protein mRNAs are natural targets of mRNA surveillance in C. elegans,"
Genes Dev, vol. 14, pp. 2173-84, 2000.
M. J. Moore, "RNA events. No end to nonsense," Science, vol. 298, pp.
370-1, 2002.
M. J. Moore, "Nuclear RNA turnover," Cell, vol. 108, pp. 431-4, 2002.
P. M. Moriarty, C. C. Reddy, and L. E. Maquat, "Selenium deficiency
reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1
by a UGA-dependent mechanism likely to be nonsense codon-mediated decay
of cytoplasmic mRNA," Mol Cell Biol, vol. 18, pp. 2932-9, 1998.
M. Morrison, K. S. Harris, and M. B. Roth, "smg mutants affect the
expression of alternatively spliced SR protein mRNAs in Caenorhabditis
elegans," Proc Natl Acad Sci U S A, vol. 94, pp. 9782-5, 1997.
O. Muhlemann, C. S. Mock-Casagrande, J. Wang, S. Li, N. Custodio, M.
Carmo-Fonseca, M. F. Wilkinson, and M. J. Moore, "Precursor RNAs harboring
nonsense codons accumulate near the site of transcription," Mol Cell, vol.
8, pp. 33-43, 2001.
D. Muhlrad and R. Parker, "Mutations affecting stability and deadenylation
of the yeast MFA2 transcript," Genes Dev, vol. 6, pp. 2100-11, 1992.
D. Muhlrad and R. Parker, "Premature translational termination triggers
mRNA decapping," Nature, vol. 370, pp. 578-81, 1994.
D. Muhlrad and R. Parker, "Recognition of yeast mRNAs as "nonsense
containing" leads to both inhibition of mRNA translation and mRNA degradation:
implications for the control of mRNA decapping," Mol Biol Cell, vol. 10,
pp. 3971-8, 1999.
D. Muhlrad and R. Parker, "Aberrant mRNAs with extended 3' UTRs are
substrates for rapid degradation by mRNA surveillance," Rna, vol. 5, pp.
1299-307, 1999.
E. Nagy and L. E. Maquat, "A rule for termination-codon position within
intron-containing genes: when nonsense affects RNA abundance," Trends Biochem
Sci, vol. 23, pp. 198-9, 1998.
E. N. Noensie and H. C. Dietz, "A strategy for disease gene identification
through nonsense-mediated mRNA decay inhibition," Nat Biotechnol, vol.
19, pp. 434-9, 2001.
M. F. Page, B. Carr, K. R. Anders, A. Grimson, and P. Anderson, "SMG-2
is a phosphorylated protein required for mRNA surveillance in Caenorhabditis
elegans and related to Upf1p of yeast," Mol Cell Biol, vol. 19, pp. 5943-51,
1999.
M. Pal, Y. Ishigaki, E. Nagy, and L. E. Maquat, "Evidence that phosphorylation
of human Upfl protein varies with intracellular location and is mediated
by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase
signaling pathway," Rna, vol. 7, pp. 5-15, 2001.
S. W. Peltz, A. H. Brown, and A. Jacobson, "mRNA destabilization triggered
by premature translational termination depends on at least three cis-acting
sequence elements and one trans-acting factor," Genes Dev, vol. 7, pp.
1737-54, 1993.
S. W. Peltz, F. He, E. Welch, and A. Jacobson, "Nonsense-mediated mRNA
decay in yeast," Prog Nucleic Acid Res Mol Biol, vol. 47, pp. 271-98, 1994.
S. W. Peltz and A. Jacobson, "mRNA stability: in trans-it," Curr Opin
Cell Biol, vol. 4, pp. 979-83, 1992.
H. A. Perlick, S. M. Medghalchi, F. A. Spencer, R. J. Kendzior, Jr.,
and H. C. Dietz, "Mammalian orthologues of a yeast regulator of nonsense
transcript stability," Proc Natl Acad Sci U S A, vol. 93, pp. 10928-32,
1996.
C. P. Ponting, "Novel eIF4G domain homologues linking mRNA translation
with nonsense-mediated mRNA decay," Trends Biochem Sci, vol. 25, pp. 423-6,
2000.
V. L. Reichert, H. Le Hir, M. S. Jurica, and M. J. Moore, "5' exon
interactions within the human spliceosome establish a framework for exon
junction complex structure and assembly," Genes Dev, vol. 16, pp. 2778-91,
2002.
L. Romao, A. Inacio, S. Santos, M. Avila, P. Faustino, P. Pacheco,
and J. Lavinha, "Nonsense mutations in the human beta-globin gene lead
to unexpected levels of cytoplasmic mRNA accumulation," Blood, vol. 96,
pp. 2895-901, 2000.
M. J. Ruiz-Echevarria, K. Czaplinski, and S. W. Peltz, "Making sense
of nonsense in yeast," Trends Biochem Sci, vol. 21, pp. 433-8, 1996.
M. J. Ruiz-Echevarria, C. I. Gonzalez, and S. W. Peltz, "Identifying
the right stop: determining how the surveillance complex recognizes and
degrades an aberrant mRNA," Embo J, vol. 17, pp. 575-89, 1998.
M. J. Ruiz-Echevarria, R. Munshi, J. Tomback, T. G. Kinzy, and S. W.
Peltz, "Characterization of a general stabilizer element that blocks deadenylation-dependent
mRNA decay," J Biol Chem, vol. 276, pp. 30995-1003, 2001.
M. J. Ruiz-Echevarria, J. M. Yasenchak, X. Han, J. D. Dinman, and S.
W. Peltz, "The upf3 protein is a component of the surveillance complex
that monitors both translation and mRNA turnover and affects viral propagation,"
Proc Natl Acad Sci U S A, vol. 95, pp. 8721-6, 1998.
G. Serin, A. Gersappe, J. D. Black, R. Aronoff, and L. E. Maquat, "Identification
and characterization of human orthologues to Saccharomyces cerevisiae Upf2
protein and Upf3 protein (Caenorhabditis elegans SMG-4)," Mol Cell Biol,
vol. 21, pp. 209-23, 2001.
R. L. Shirley, A. S. Ford, M. R. Richards, M. Albertini, and M. R.
Culbertson, "Nuclear import of Upf3p is mediated by importin-alpha/-beta
and export to the cytoplasm is required for a functional nonsense-mediated
mRNA decay pathway in yeast," Genetics, vol. 161, pp. 1465-82, 2002.
R. L. Shirley, M. J. Lelivelt, L. R. Schenkman, J. N. Dahlseid, and
M. R. Culbertson, "A factor required for nonsense-mediated mRNA decay in
yeast is exported from the nucleus to the cytoplasm by a nuclear export
signal sequence," J Cell Sci, vol. 111 ( Pt 21), pp. 3129-43, 1998.
A. B. Shyu and M. F. Wilkinson, "The double lives of shuttling mRNA
binding proteins," Cell, vol. 102, pp. 135-8, 2000.
L. S. Stephenson and L. E. Maquat, "Cytoplasmic mRNA for human triosephosphate
isomerase is immune to nonsense-mediated decay despite forming polysomes,"
Biochimie, vol. 78, pp. 1043-7, 1996.
X. Sun, X. Li, P. M. Moriarty, T. Henics, J. P. LaDuca, and L. E. Maquat,
"Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide
glutathione peroxidase is detectable in cultured cells but masked or inhibited
in rat tissues," Mol Biol Cell, vol. 12, pp. 1009-17, 2001.
X. Sun and L. E. Maquat, "mRNA surveillance in mammalian cells: the
relationship between introns and translation termination," Rna, vol. 6,
pp. 1-8, 2000.
X. Sun and L. E. Maquat, "Nonsense-mediated decay: assaying for effects
on selenoprotein mRNAs," Methods Enzymol, vol. 347, pp. 49-57, 2002.
X. Sun, P. M. Moriarty, and L. E. Maquat, "Nonsense-mediated decay
of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position,"
Embo J, vol. 19, pp. 4734-44, 2000.
X. Sun, H. A. Perlick, H. C. Dietz, and L. E. Maquat, "A mutated human
homologue to yeast Upf1 protein has a dominant-negative effect on the decay
of nonsense-containing mRNAs in mammalian cells," Proc Natl Acad Sci U
S A, vol. 95, pp. 10009-14, 1998.
A. Sureau, R. Gattoni, Y. Dooghe, J. Stevenin, and J. Soret, "SC35
autoregulates its expression by promoting splicing events that destabilize
its mRNAs," Embo J, vol. 20, pp. 1785-96, 2001.
C. Valentin, M. Cohen-Solal, L. Maquat, M. Horanyi, M. Inselt-Kovacs,
and S. Hollan, "Identical germ-line mutations in the triosephosphate isomerase
alleles of two brothers are associated with distinct clinical phenotypes,"
C R Acad Sci III, vol. 323, pp. 245-50, 2000.
S. Vasudevan and S. W. Peltz, "Regulated ARE-mediated mRNA decay in
Saccharomyces cerevisiae," Mol Cell, vol. 7, pp. 1191-200, 2001.
E. Wagner and J. Lykke-Andersen, "mRNA surveillance: the perfect persist,"
J Cell Sci, vol. 115, pp. 3033-8, 2002.
J. Wang, Y. F. Chang, J. I. Hamilton, and M. F. Wilkinson, "Nonsense-associated
altered splicing: a frame-dependent response distinct from nonsense-mediated
decay," Mol Cell, vol. 10, pp. 951-7, 2002.
J. Wang, J. P. Gudikote, O. R. Olivas, and M. F. Wilkinson, "Boundary-independent
polar nonsense-mediated decay," EMBO Rep, vol. 3, pp. 274-9, 2002.
J. Wang, V. M. Vock, S. Li, O. R. Olivas, and M. F. Wilkinson, "A quality
control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts
by a mechanism requiring UPF2 and translation," J Biol Chem, vol. 277,
pp. 18489-93, 2002.
W. Wang, K. Czaplinski, Y. Rao, and S. W. Peltz, "The role of Upf proteins
in modulating the translation read-through of nonsense-containing transcripts,"
Embo J, vol. 20, pp. 880-90, 2001.
Y. Weng, K. Czaplinski, and S. W. Peltz, "Genetic and biochemical characterization
of mutations in the ATPase and helicase regions of the Upf1 protein," Mol
Cell Biol, vol. 16, pp. 5477-90, 1996.
Y. Weng, K. Czaplinski, and S. W. Peltz, "Identification and characterization
of mutations in the UPF1 gene that affect nonsense suppression and the
formation of the Upf protein complex but not mRNA turnover," Mol Cell Biol,
vol. 16, pp. 5491-506, 1996.
Y. Weng, K. Czaplinski, and S. W. Peltz, "ATP is a cofactor of the
Upf1 protein that modulates its translation termination and RNA binding
activities," Rna, vol. 4, pp. 205-14, 1998.
M. F. Wilkinson and A. B. Shyu, "Multifunctional regulatory proteins
that control gene expression in both the nucleus and the cytoplasm," Bioessays,
vol. 23, pp. 775-87, 2001.
M. F. Wilkinson and A. B. Shyu, "RNA surveillance by nuclear scanning?,"
Nat Cell Biol, vol. 4, pp. E144-7, 2002.
G. M. Wilson, Y. Sun, J. Sellers, H. Lu, N. Penkar, G. Dillard, and
G. Brewer, "Regulation of AUF1 expression via conserved alternatively spliced
elements in the 3' untranslated region," Mol Cell Biol, vol. 19, pp. 4056-64,
1999.
C. J. Wilusz, W. Wang, and S. W. Peltz, "Curbing the nonsense: the
activation and regulation of mRNA surveillance," Genes Dev, vol. 15, pp.
2781-5, 2001.
C. J. Wilusz, M. Wormington, and S. W. Peltz, "The cap-to-tail guide
to mRNA turnover," Nat Rev Mol Cell Biol, vol. 2, pp. 237-46, 2001.
A. Yamashita, T. Ohnishi, I. Kashima, Y. Taya, and S. Ohno, "Human
SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates
with components of the mRNA surveillance complex and is involved in the
regulation of nonsense-mediated mRNA decay," Genes Dev, vol. 15, pp. 2215-28,
2001.
J. Zhang and L. E. Maquat, "Evidence that the decay of nucleus-associated
nonsense mRNA for human triosephosphate isomerase involves nonsense codon
recognition after splicing," Rna, vol. 2, pp. 235-43, 1996.
J. Zhang and L. E. Maquat, "Evidence that translation reinitiation
abrogates nonsense-mediated mRNA decay in mammalian cells," Embo J, vol.
16, pp. 826-33, 1997.
J. Zhang, X. Sun, Y. Qian, J. P. LaDuca, and L. E. Maquat, "At least
one intron is required for the nonsense-mediated decay of triosephosphate
isomerase mRNA: a possible link between nuclear splicing and cytoplasmic
translation," Mol Cell Biol, vol. 18, pp. 5272-83, 1998.
J. Zhang, X. Sun, Y. Qian, and L. E. Maquat, "Intron function in the
nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA
splicing in the nucleus can influence mRNA translation in the cytoplasm,"
Rna, vol. 4, pp. 801-15, 1998.
S. Zhang, M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz, "Identification
and characterization of a sequence motif involved in nonsense-mediated
mRNA decay," Mol Cell Biol, vol. 15, pp. 2231-44, 1995.
S. Zhang, E. M. Welch, K. Hogan, A. H. Brown, S. W. Peltz, and A. Jacobson,
"Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA
decay pathway in Saccharomyces cerevisiae," Rna, vol. 3, pp. 234-44, 1997.
S. Zhang, C. J. Williams, K. Hagan, and S. W. Peltz, "Mutations in
VPS16 and MRT1 stabilize mRNAs by activating an inhibitor of the decapping
enzyme," Mol Cell Biol, vol. 19, pp. 7568-76, 1999.
S. Zhang, C. J. Williams, M. Wormington, A. Stevens, and S. W. Peltz,
"Monitoring mRNA decapping activity," Methods, vol. 17, pp. 46-51, 1999.
S. Zhu, W. Li, and Z. Cao, "A naturally occurring non-coding fusion
transcript derived from scorpion venom gland: implication for the regulation
of scorpion toxin gene expression," FEBS Lett, vol. 508, pp. 241-4, 2001.